Many repair processes exist to ensure genetic stability, such as nucleotide excision repair (NER), homologous recombination (HR), and non-homologous end-joining (NHEJ). However, deregulations can occur in these pathways and often lead to disease development like cancer or neurodegenerative disorders. In order to successfully develop new therapies, it is crucial to research DNA repair processes to achieve a full understanding of their mechanism. As DNA repair is a highly complex and dynamic process that involves numerous different proteins, to gain the necessary insights, scientists need to observe the interplay of the components in real-time and at the molecular level.
C-Trap® Optical Tweezers – Fluorescence & Label-free Microscopy provides exactly these capabilities, empowering your research to gather valuable insights into the detailed mechanisms governing DNA repair.
Visualization of DNA-binding proteins and their interactions with DNA in real-time
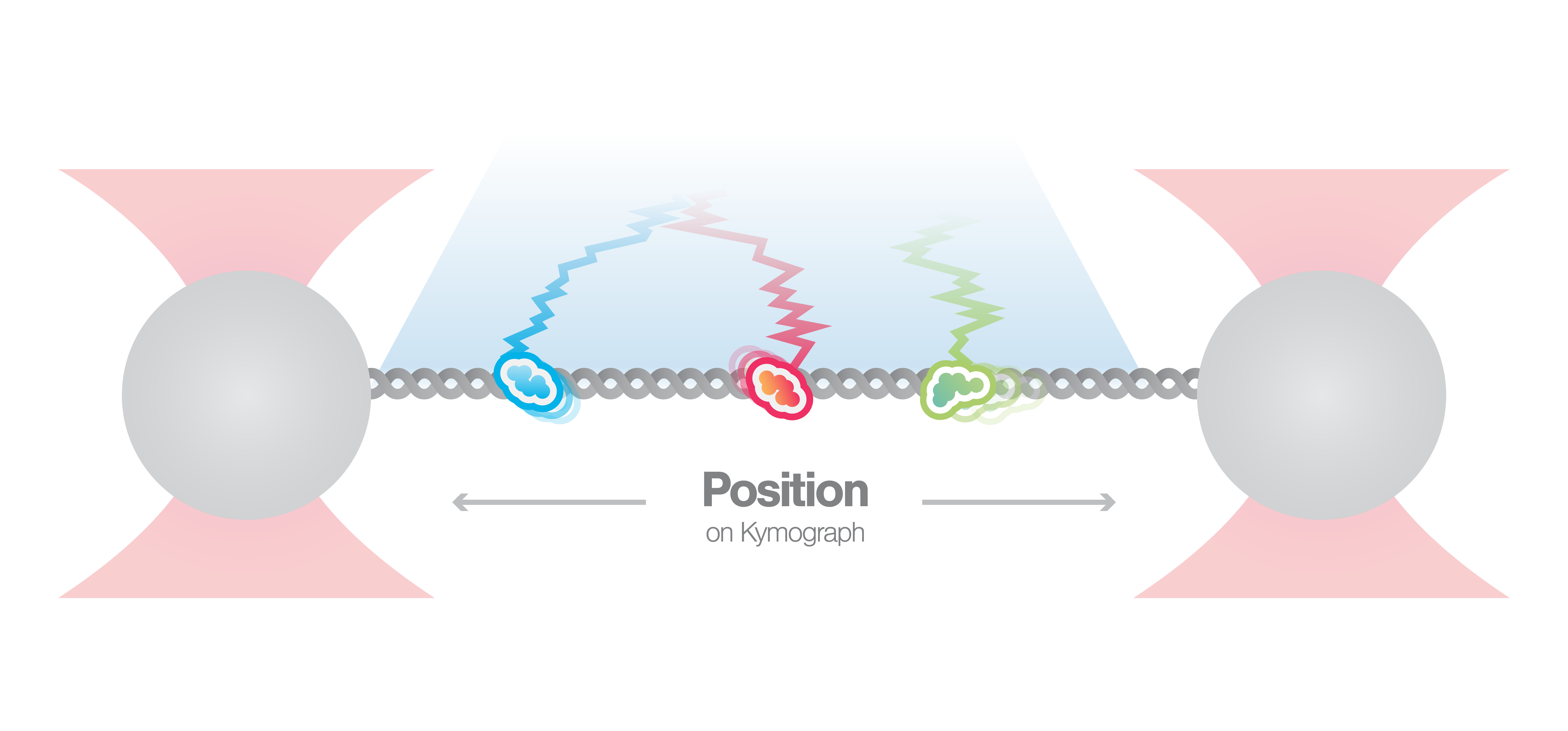
By utilizing the fully correlated integration between multi-color fluorescence microscopy and optical tweezers, the C-Trap enables observing the dynamics of multiple different proteins at the same time. In this experiment, the dynamics of two different DNA-binding repair proteins are visualized and presented in a kymograph. This unique form of visualization allows to track their movements and analyze their interaction with DNA as well as among each other.
Double-stranded breaks are often repaired by the NHEJ pathway. Hereby, for the ligation step of DNA ends the DNA ligase is interacting with protein XRCC4 which forms complexes with XRCC4-like factor (XLF) to bridge DNA. In this experiment, a single double-stranded DNA is tethered between two beads, and the interaction of fluorescently-labeled XRCC4 and XLF is observed in real-time.
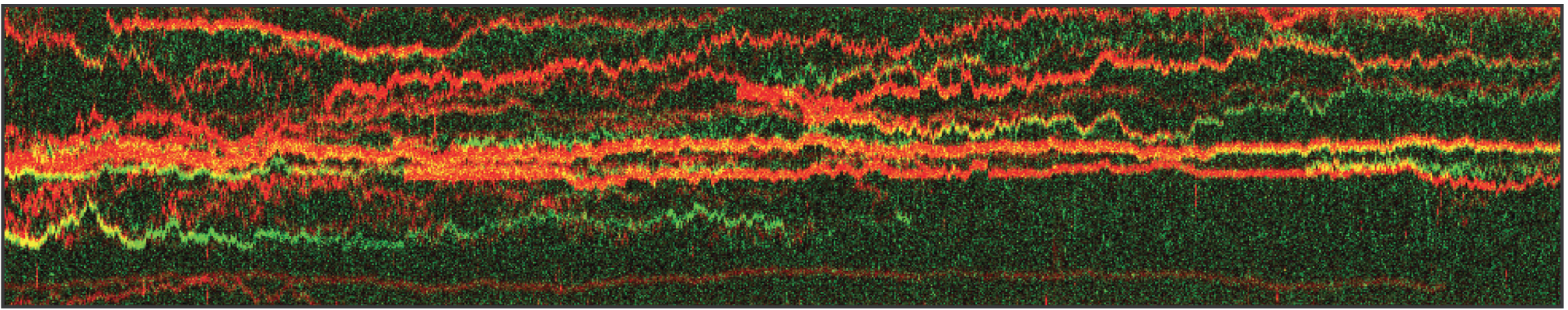
Figure 1 shows the position of XRCC4 and XLF on DNA over time. The data allow not only to visualize the dynamics but also to quantify the incidence of the protein forms. Here, N=94 events were detected whereby 9% of XRCC4 alone, 62% of XLF alone, and 29% of XRCC4-XLF complexes. Using the C-Trap to record kymographs of multiple fluorescently-labeled components gives real-time insights into DNA-protein and protein-protein interactions involved in DNA repair simultaneously.
1 Kymograph showing the dynamics of fluorescently-labeled repair proteins XRCC4 (green), XLF (red), and XRCC4-XLF complexes (yellow) on optically-trapped DNA. Protein concentration is 5 nM.
Application note | STED
Brouwer et al. (2016) Nature
Visualization and quantification of RAD-51 nucleation on single-stranded DNA under different conditions
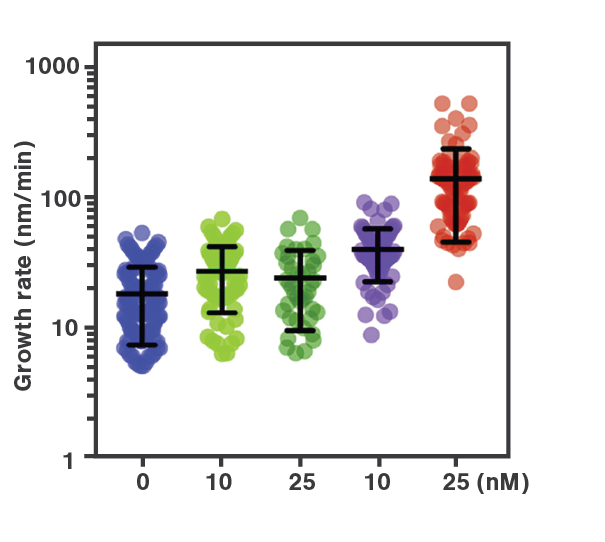
The combination of microfluidics, optical tweezers, and fluorescence microscopy within the C-Trap enables the users to easily observe the interactions of DNA-binding proteins under different conditions. Using these features, scientists from the labs of Prof. Rueda and Prof. Simon Boulton analyzed the assembly of RAD51 filaments with and without the presence of different mediators – factors that are important for homologous recombination (HR) during DNA double-strand break repair. The data collected by the correlated fluorescence imaging were used to quantify and plot the dynamics of the observed interactions.
During HR, RPA proteins coat resected DNA ends and are then replaced by RAD-51 which forms long nucleoprotein filaments. Current studies implicate that BRCA2 and RAD-51 paralogs contribute to RAD-51 filament formation but the understanding of how these mediators affect the filament assembly and growth has been so far unclear. Here, real-time single-molecule imaging using the C-Trap was used to investigate RPA displacement and the sequential assembly of RAD-51 protein filaments.
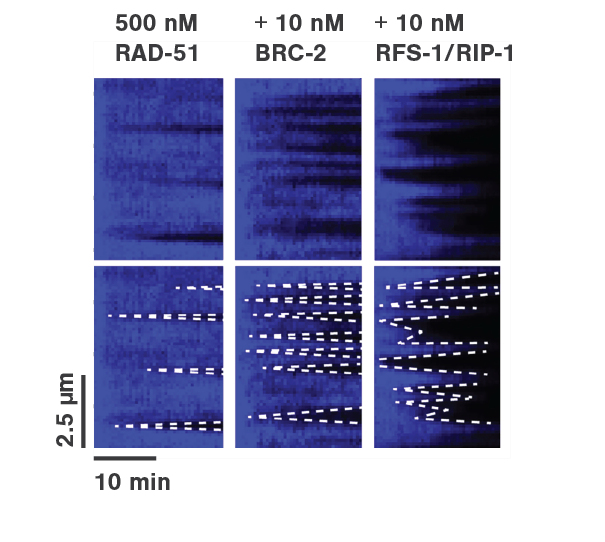
To mimic the HR process, optically-trapped double-stranded DNA was melted using force and the produced single-stranded ends were coated with eGFP-labeled RPA. Then the optical traps were moved to another channel of the microfluidics chamber containing either RAD-51 alone or in presence of BRC-2 or RFS-1/RPI-1. The loss of eGFP fluorescence (as shown in Figure 2a) represented the replacement of RPA-eGFP by RAD-51. The growth rates of individual RAD-51 filaments were clearly stimulated by the presence of BRC-2 as well as RFS-1/RIP-1 (Figure 2b). Based on their results, the scientists suggested that BRC-2 primarily acts as RAD-51 nucleation factor whereas RFS-1/RIP-1 acts as a filament growth factor.
Employing the C-Trap enabled the scientists to recreate homologous recombination in situ, visualize individual protein dynamics, and gather direct proof of the ongoing molecular interactions during this complex process.
Application note | DNA repair
Force extension and manipulation of DNA-protein interactions
Optical tweezers provide a highly versatile system that allows manipulation of single molecules at the nanoscale and the read-out of valuable data. Using force and extension measurements, the influence of repair proteins on DNA structure and physical properties can be analyzed.
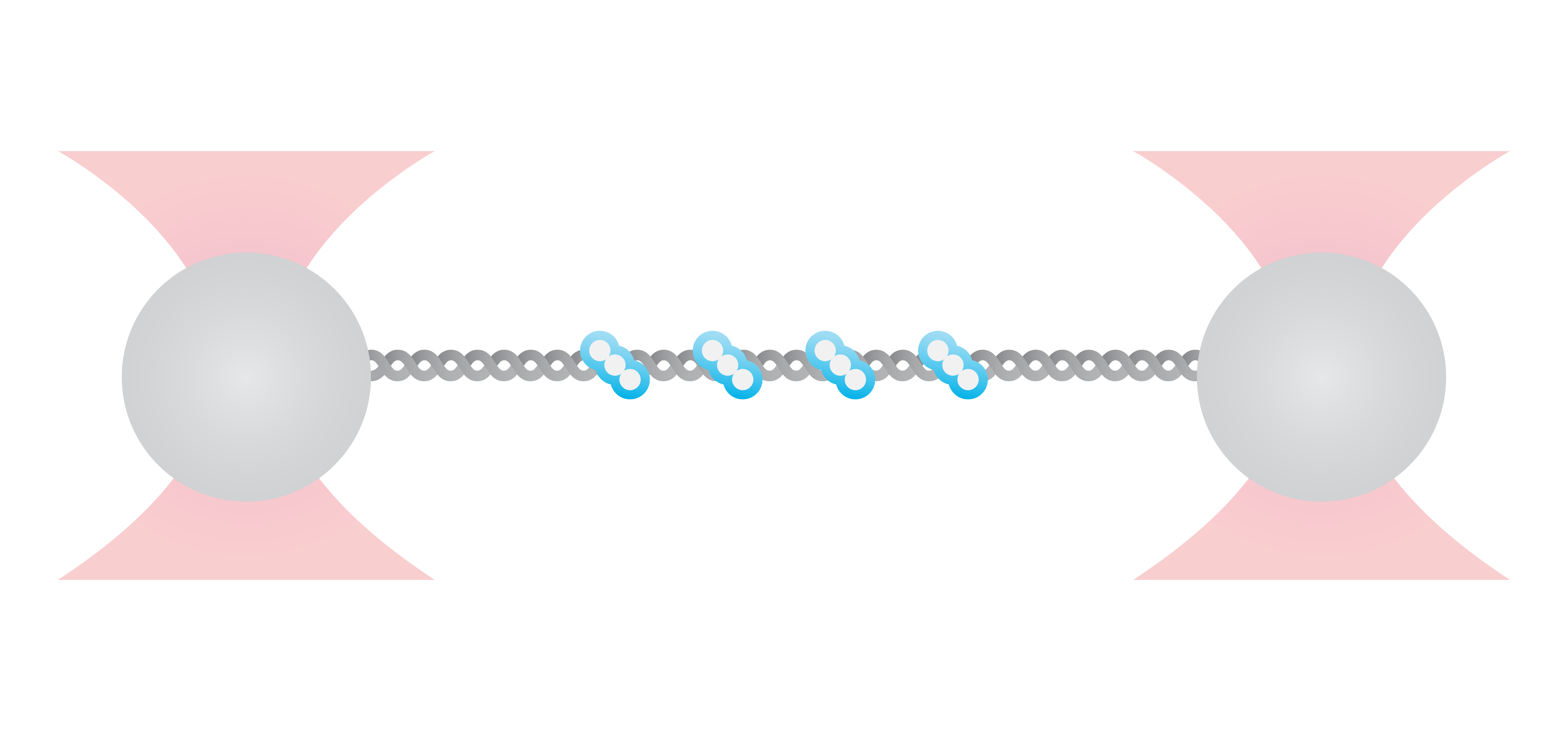
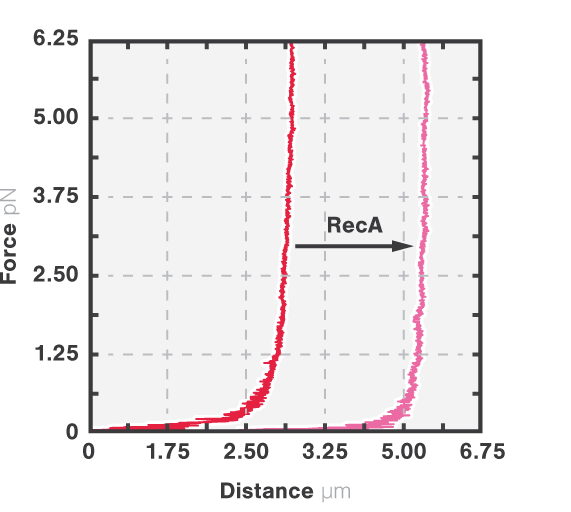
RecA is a bacterial DNA repair protein that fulfills different activities in HR. It forms helical filaments around the DNA molecule and promotes DNA synapsis, heteroduplex formation, and the exchange between homologous DNA strands. To understand how RecA facilitates these functions, the effect of RecA-DNA assembly on the mechanical properties of DNA was analyzed using optical tweezers.
Here, DNA tethered to optically trapped beads was stretched while measuring the force and extension before and after being coated with RecA. With RecA, a clear shift in the force-distance curve occurred (Figure 3, arrow), showing that less force is necessary to unravel the DNA-RecA complex. This suggests an increased stiffness of DNA: the RecA filaments prevent it from coiling thus keeping the bases accessible for HR processes like heteroduplex formation and strand exchange.
Employing optical tweezers – m-Trap or C-Trap – enables scientists to extract information on how different repair proteins influence the mechanical properties of DNA. Noteworthy: A combination with simultaneous fluorescence measurements – available with the C-Trap system – would allow correlating the physical properties with the binding location and quantity of DNA repair proteins.
Brouwer et al. (2017) Cell Reports